I'm trying to match the histograms of two images (in MATLAB this could be done using imhistmatch). Is there an equivalent function available from a standard Python library? I've looked at OpenCV, scipy, and numpy but don't see any similar functionality.
I previously wrote an answer here explaining how to do piecewise linear interpolation on an image histogram in order to enforce particular ratios of highlights/midtones/shadows.
The same basic principles underlie histogram matching between two images. Essentially you compute the cumulative histograms for your source and template images, then interpolate linearly to find the unique pixel values in the template image that most closely match the quantiles of the unique pixel values in the source image:
import numpy as np
def hist_match(source, template):
"""
Adjust the pixel values of a grayscale image such that its histogram
matches that of a target image
Arguments:
-----------
source: np.ndarray
Image to transform; the histogram is computed over the flattened
array
template: np.ndarray
Template image; can have different dimensions to source
Returns:
-----------
matched: np.ndarray
The transformed output image
"""
oldshape = source.shape
source = source.ravel()
template = template.ravel()
# get the set of unique pixel values and their corresponding indices and
# counts
s_values, bin_idx, s_counts = np.unique(source, return_inverse=True,
return_counts=True)
t_values, t_counts = np.unique(template, return_counts=True)
# take the cumsum of the counts and normalize by the number of pixels to
# get the empirical cumulative distribution functions for the source and
# template images (maps pixel value --> quantile)
s_quantiles = np.cumsum(s_counts).astype(np.float64)
s_quantiles /= s_quantiles[-1]
t_quantiles = np.cumsum(t_counts).astype(np.float64)
t_quantiles /= t_quantiles[-1]
# interpolate linearly to find the pixel values in the template image
# that correspond most closely to the quantiles in the source image
interp_t_values = np.interp(s_quantiles, t_quantiles, t_values)
return interp_t_values[bin_idx].reshape(oldshape)
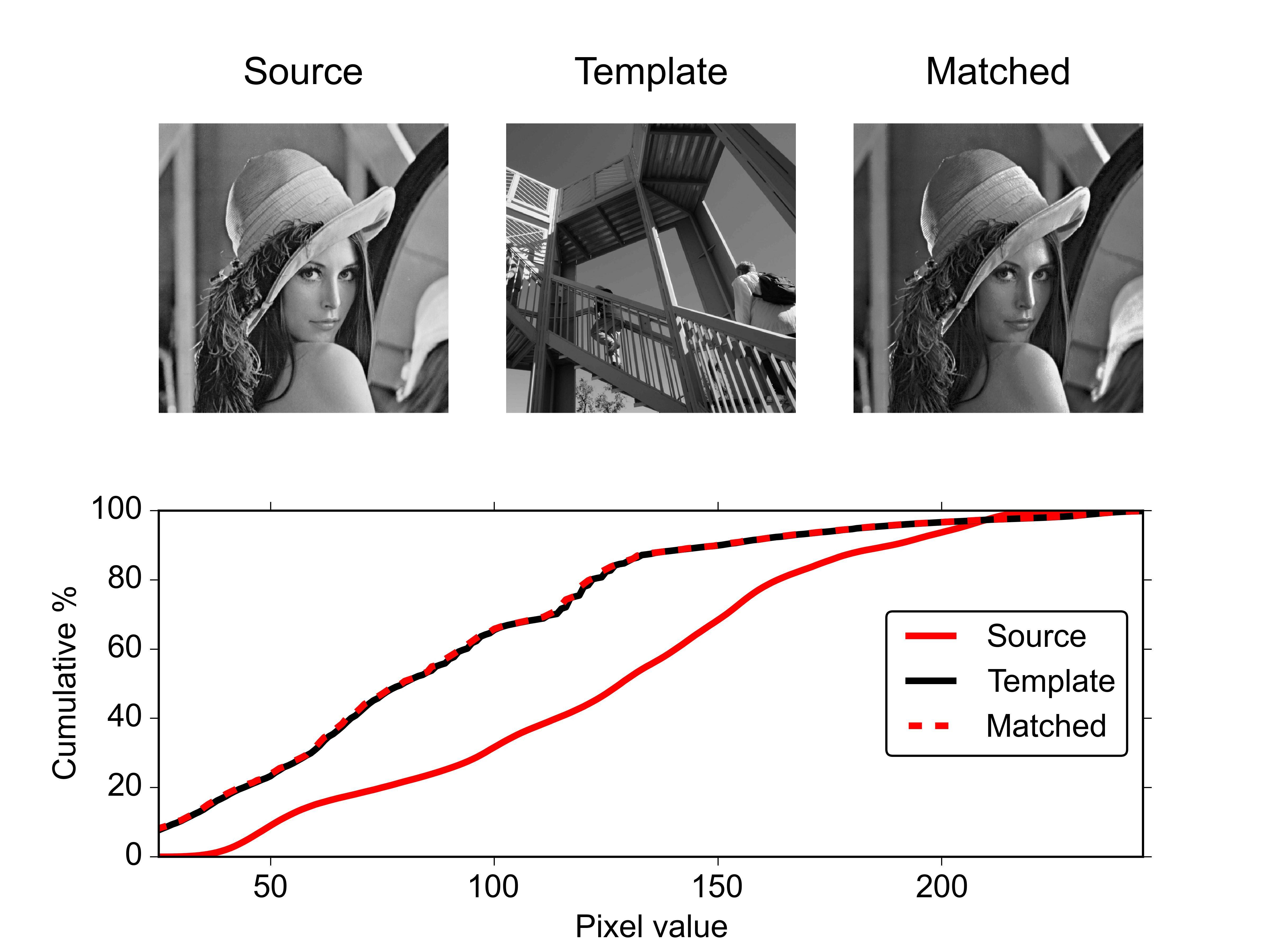
For example:
from matplotlib import pyplot as plt
from scipy.misc import lena, ascent
source = lena()
template = ascent()
matched = hist_match(source, template)
def ecdf(x):
"""convenience function for computing the empirical CDF"""
vals, counts = np.unique(x, return_counts=True)
ecdf = np.cumsum(counts).astype(np.float64)
ecdf /= ecdf[-1]
return vals, ecdf
x1, y1 = ecdf(source.ravel())
x2, y2 = ecdf(template.ravel())
x3, y3 = ecdf(matched.ravel())
fig = plt.figure()
gs = plt.GridSpec(2, 3)
ax1 = fig.add_subplot(gs[0, 0])
ax2 = fig.add_subplot(gs[0, 1], sharex=ax1, sharey=ax1)
ax3 = fig.add_subplot(gs[0, 2], sharex=ax1, sharey=ax1)
ax4 = fig.add_subplot(gs[1, :])
for aa in (ax1, ax2, ax3):
aa.set_axis_off()
ax1.imshow(source, cmap=plt.cm.gray)
ax1.set_title('Source')
ax2.imshow(template, cmap=plt.cm.gray)
ax2.set_title('template')
ax3.imshow(matched, cmap=plt.cm.gray)
ax3.set_title('Matched')
ax4.plot(x1, y1 * 100, '-r', lw=3, label='Source')
ax4.plot(x2, y2 * 100, '-k', lw=3, label='Template')
ax4.plot(x3, y3 * 100, '--r', lw=3, label='Matched')
ax4.set_xlim(x1[0], x1[-1])
ax4.set_xlabel('Pixel value')
ax4.set_ylabel('Cumulative %')
ax4.legend(loc=5)
For a pair of RGB images you could apply this function separately to each color channel.
Here is yet another implementation based on this and scikit-image exposure's cumulative_distribution function which uses np.interp similar to ali_m's implementation. The input and template image is assumed to be gray-scale and with pixel values as integers in [0,255].
from skimage.exposure import cumulative_distribution
import matplotlib.pylab as plt
import numpy as np
def cdf(im):
'''
computes the CDF of an image im as 2D numpy ndarray
'''
c, b = cumulative_distribution(im)
# pad the beginning and ending pixels and their CDF values
c = np.insert(c, 0, [0]*b[0])
c = np.append(c, [1]*(255-b[-1]))
return c
def hist_matching(c, c_t, im):
'''
c: CDF of input image computed with the function cdf()
c_t: CDF of template image computed with the function cdf()
im: input image as 2D numpy ndarray
returns the modified pixel values
'''
pixels = np.arange(256)
# find closest pixel-matches corresponding to the CDF of the input image, given the value of the CDF H of
# the template image at the corresponding pixels, s.t. c_t = H(pixels) <=> pixels = H-1(c_t)
new_pixels = np.interp(c, c_t, pixels)
im = (np.reshape(new_pixels[im.ravel()], im.shape)).astype(np.uint8)
return im
The output is shown below:
I would like to add a minor addition to both the solution written above. If somebody plans to make this as a global function (such as for grayscale images), it would be a good idea to convert the final matched array into its corresponding format (numpy.uint8). This might help in future image conversions without creating conflicts.
def hist_norm(source, template):
olddtype = source.dtype
oldshape = source.shape
source = source.ravel()
template = template.ravel()
s_values, bin_idx, s_counts = np.unique(source, return_inverse=True,
return_counts=True)
t_values, t_counts = np.unique(template, return_counts=True)
s_quantiles = np.cumsum(s_counts).astype(np.float64)
s_quantiles /= s_quantiles[-1]
t_quantiles = np.cumsum(t_counts).astype(np.float64)
t_quantiles /= t_quantiles[-1]
interp_t_values = np.interp(s_quantiles, t_quantiles, t_values)
interp_t_values = interp_t_values.astype(olddtype)
return interp_t_values[bin_idx].reshape(oldshape)
来源:https://stackoverflow.com/questions/32655686/histogram-matching-of-two-images-in-python-2-x